The participation on the COVID-19 Genomic Surveillance Regional Network is open to all the countries of the Americas through the National Public Health Laboratories.
For more detailed information on partnering with us, please contact the PAHO Regional Office: leitejul@paho.org, ricoj@paho.org.
SARS-CoV-2 Genomic Sequencing Laboratories
© Pan American Health Organization, 2020. All rights reserved.
Report production: Paho Health Emergencies Department (PHE): Influenza Surveillance Team/ Infectious Hazard Management (IHM) and Data Management, Analytics & Products/ Health Emergency Information & Risk Assessments (HIM).
Disclaimer: The designations employed and the presentation of the material in these maps do not imply the expression of any opinion whatsoever on the part of the Secretariat of the Pan American Health Organization concerning the legal status of any country, territory, city or area or of its authorities, or concerning the delimitation of its frontiers or boundaries. Dotted and dashed lines on maps represent approximate border lines for which there may not yet be full agreement.
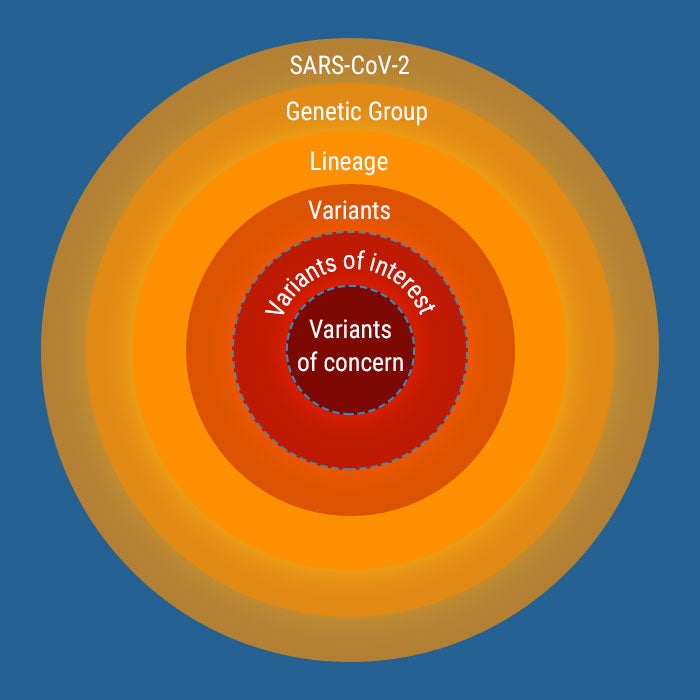
Variants of SARS-CoV-2 in the Americas
Since the initial genomic characterization of SARS-CoV-2, the virus has been divided into different genetic groups. The occurrence of mutations is a natural and expected event within the evolution process of the virus.
- Mutation refers to any change in the virus genome.
- Some specific mutations define the viral genetic groups currently circulating globally.
- In general, a mutated virus is considered a variant of the original virus.
- There are different types of mutations. Variants can differ by one or more or those.
- Although most of the mutations have no impact, some may result in the virus being more transmissible, or facilitating the virus to escape immune response.
- In the community, the higher the transmission level, the higher is the likelihood of viral mutations to occur.
When these variants have a potential impact or risk for public health, they are considered variants of concern (VOC).
Variants classification1
A SARS-CoV-2 isolate is a variant of interest (VOI) if it is phenotypically changed compared to a reference isolate or has a genome with mutations that lead to amino acid changes associated with established or suspected phenotypic implications1;
AND
has been identified to cause community transmission2/multiple COVID-19 cases/clusters, or has been detected in multiple countries;
OR
is otherwise assessed to be a VOI by WHO in consultation with the WHO SARS-CoV-2 Virus Evolution Working Group.
A VOI (as defined above) is a variant of concern (VOC) if, through a comparative assessment, it has been demonstrated to be associated with
- Increase in transmissibility or detrimental change in COVID-19 epidemiology;
- Increase in virulence or change in clinical disease presentation; or
- Decrease in effectiveness of public health and social measures or available diagnostics, vaccines, therapeutics.
OR
assessed to be a VOC by WHO in consultation with the WHO SARS-CoV-2 Virus Evolution Working Group.
1https://www.who.int/publications/m/item/covid-19-weekly-epidemiological-update
Monitoring of SARS-CoV-2 variants
Virus mutations or variants are being monitored from the start of the COVID-19 pandemic through the Global Initiative on Sharing Avian Influenza Data (GISAID) sequencing database. WHO routinely assesses if variants of SARSCoV-2 have an impact on:
- Virus transmissibility;
- Disease severity;
- Efficacy of diagnostics, therapeutics and vaccines.
Risk assessment for variants of concern to determine if there will be public health implications are routinely performed.
Main actions by a Member State, if a VOI or VOC is identified:
- Immediately report to PAHO/WHO initial cases/clusters associated with VOI or VOC infection through the IHR mechanism.
- Submit complete genome sequences and associated metadata to a publicly available database, such as GISAID.
- Where capacity exists and in coordination with the international community, perform field investigations to improve understanding of the potential impacts of the VOI or VOC on COVID-19 epidemiology, severity, effectiveness of public health and social measures, or other relevant characteristics.
- Perform laboratory assessments or contact WHO for support to conduct laboratory assessments on the impact of the VOI or VOC on diagnostic methods, immune responses, antibody neutralization or other relevant characteristics.
SARS CoV2 Genomic Suerveillance Documents
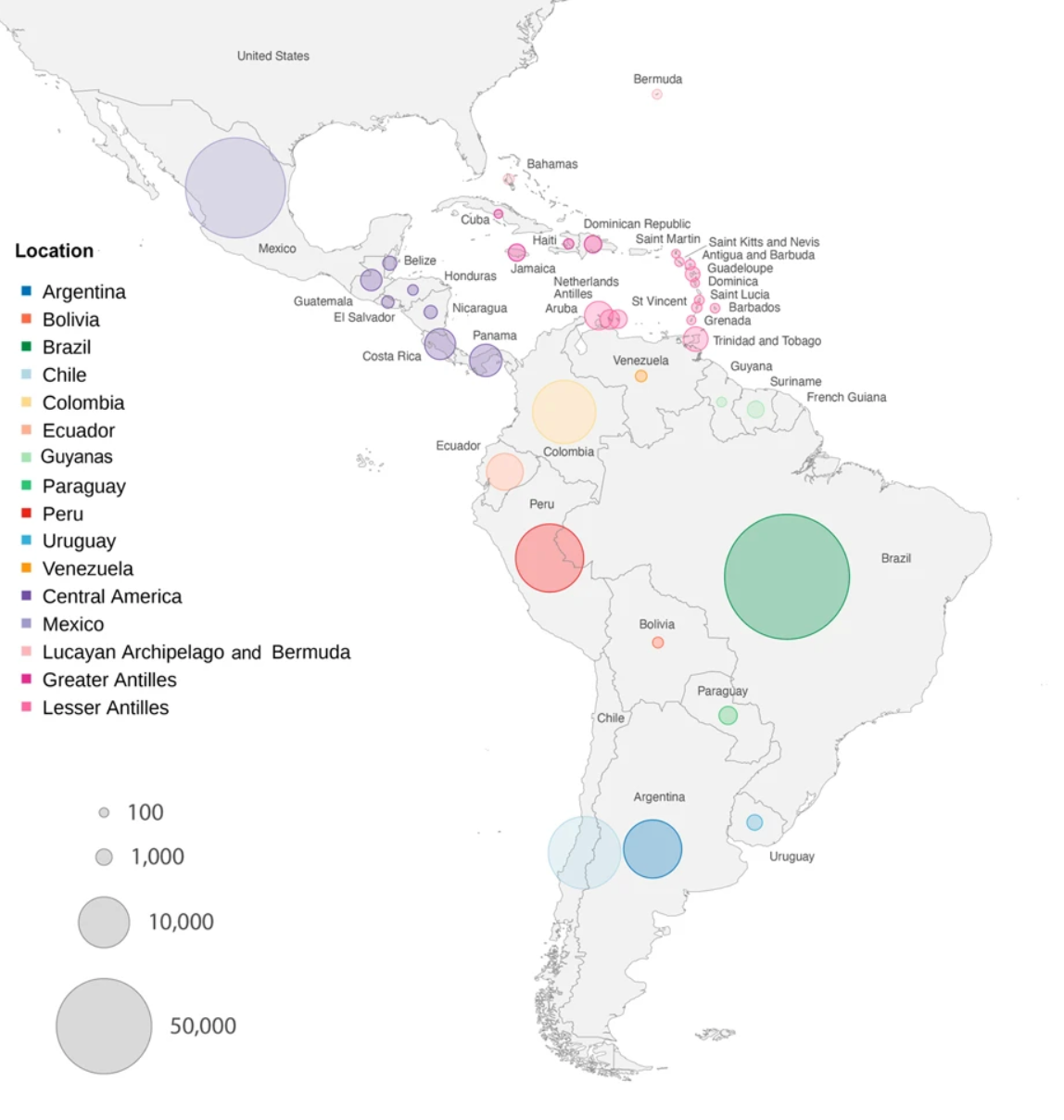
Dispersion patterns of SARS-CoV-2 variants Gamma, Lambda and Mu in Latin America and the Caribbean
Published: 28 February 2024 - Nature Communications
Latin America and Caribbean (LAC) regions were an important epicenter of the COVID-19 pandemic and SARS-CoV-2 evolution. Through the COVID-19 Genomic Surveillance Regional Network (COVIGEN), LAC countries produced an important number of genomic sequencing data that made possible an enhanced SARS-CoV-2 genomic surveillance capacity in the Americas, paving the way for characterization of emerging variants and helping to guide the public health response. In this study we analyzed approximately 300,000 SARS-CoV-2 sequences generated between February 2020 and March 2022 by multiple genomic surveillance efforts in LAC and reconstructed the diffusion patterns of the main variants of concern (VOCs) and of interest (VOIs) possibly originated in the Region. Our phylogenetic analysis revealed that the spread of variants Gamma, Lambda and Mu reflects human mobility patterns due to variations of international air passenger transportation and gradual lifting of social distance measures previously implemented in countries. Our results highlight the potential of genetic data to reconstruct viral spread and unveil preferential routes of viral migrations that are shaped by human mobility patterns.